Epidemics, evolution, and too much math
Computational methods for forecasting viral evolution
Marlin Figgins
2022-09-08
Who am I?
- UChicago alumnus (BS, Mathematics 2019)
- Current PhD student in Applied Math at Fred Hutch and the University of Washington in the Bedford lab.
- Now, I study transmission and evolution of pathogens like influenza and SARS-CoV-2.
Epidemics are often observed with counts
We use \(R_{t}\) to infer the direction of epidemics
{.fig-align=“center”}
Pathogens have an evolutionary history
Mutation can lead to transmission differences
Variant differences may not show in cases
Variant frequencies allow us to monitor change
Turning frequencies into growth advantages
Transmission models with various strains
{.fig-align=“center”}
Applying this to SARS-CoV-2 and COVID-19 in the USA
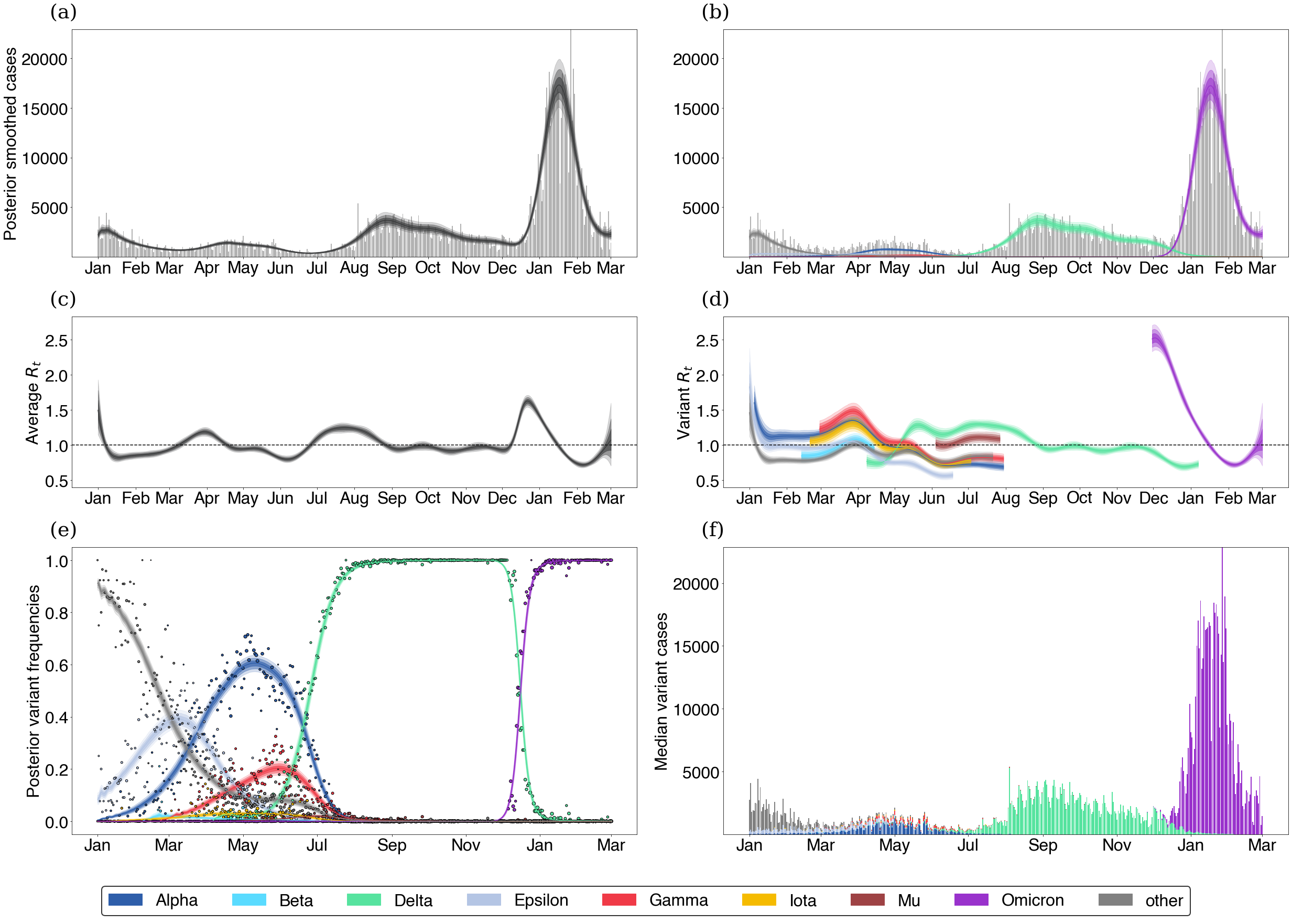
Applying this to SARS-CoV-2 and COVID-19 in the USA
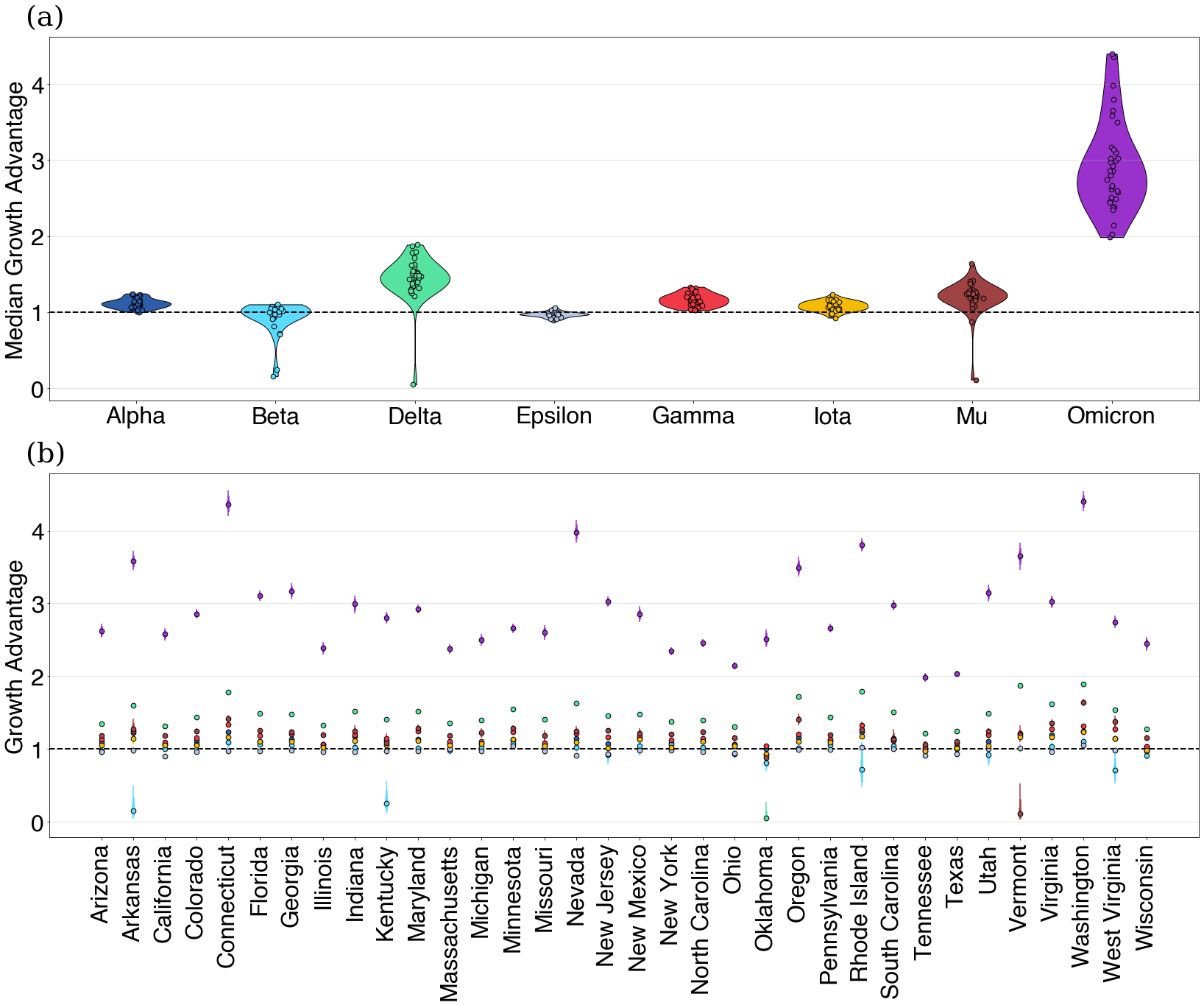
Applying this to SARS-CoV-2 and COVID-19 in the USA
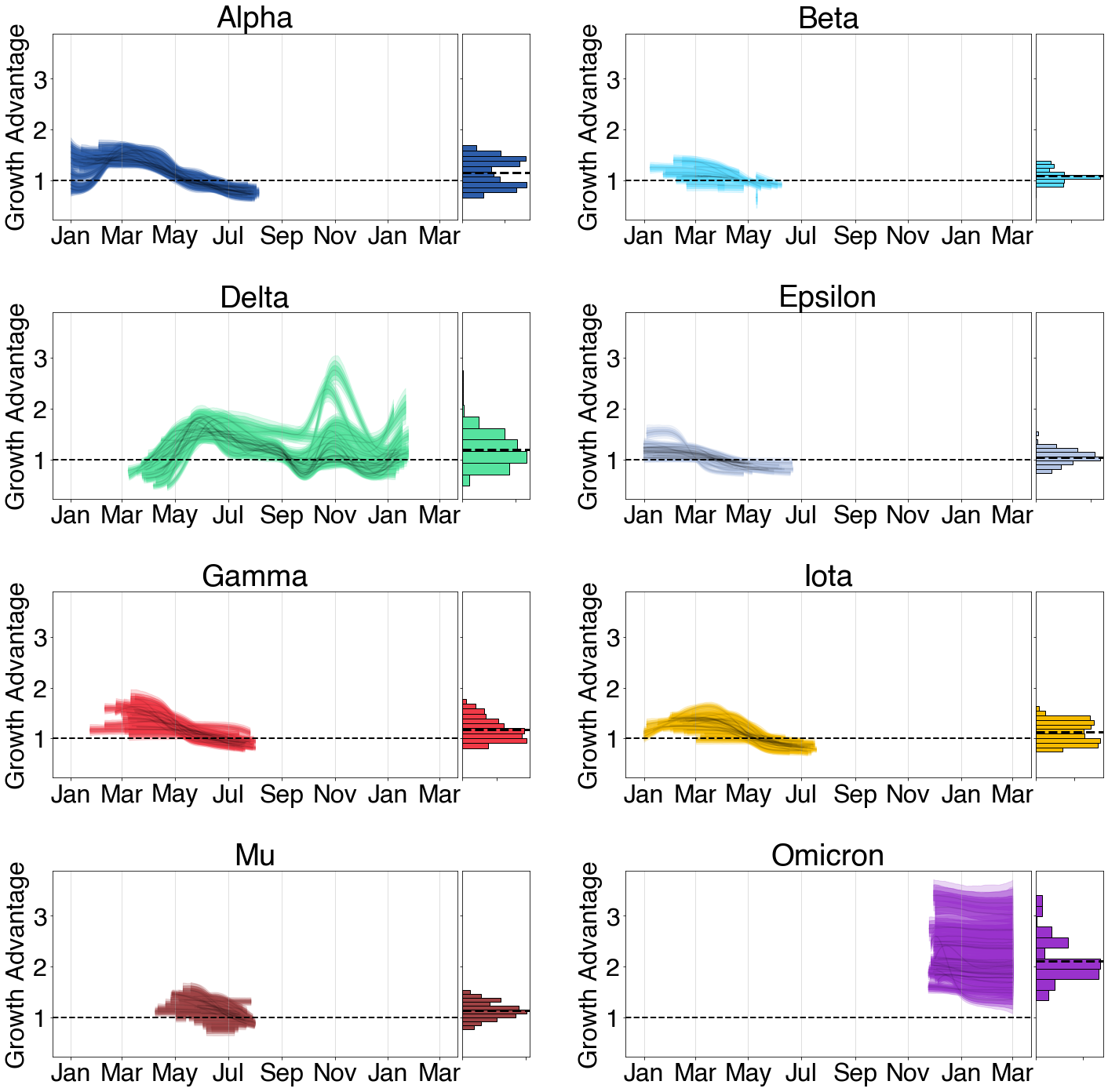
Developing tools for analyzing epidemics
- How did we go from a one-off model to a tool that we can continually use?
- My research is focused on building tools and toolkits for analyzing evolution of pathogens.
- This includes both software (like that used to make today’s figures) and mathematical machinery that teaches how and why growth advantages may appear between pathogen variants.
Takeaways
- Understanding epidemics is complicated by feedback loop between evolution and transmission
- Mathematics and statistics are tools for analyzing data that can provide new insight.
- Once we have an idea in mind, we can build re-usable and customizable tools for analyzing new data to make new analyses easier.
- This allows us to make many forecasts as we receive new data and opens the door for evolutionary forecasting